Prevalence_of_COVID-19_in_BaWu__.pdf
Dieses Dokument ist Teil der Anfrage „Zwischenergebnis der sog. Corona-Kinder-Studie“
1 Prevalence of COVID-19 in children in Baden-Württemberg Preliminary study report Klaus-Michael Debatin, University Children’s Hospital Ulm Philipp Henneke, University Children’s Hospital Freiburg Georg Friedrich Hoffmann, University Children’s Hospital Heidelberg Hans-Georg Kräusslich, Department of Infectious Diseases, Heidelberg University Hospital Hanna Renk, University Children’s Hospital Tübingen on behalf of all members of the study group This study was supported by a grant from the State of Baden-Württemberg

2 Summary 1. Study Population: This is an interim analysis not included in the initial study protocol performed on 4,932 subjects (2,466 children and 2,466 corresponding parents) with some minor data cleaning and validation still ongoing. 2. Detection of acute SARS-CoV-2 infection by RT-PCR: During the sampling nd th period between April 22 and May 15 , 2020, among the 4,932 individuals who were tested by RT-PCR, only 2 subjects (0.04%), one child and the corresponding parent, were positive for SARS-CoV-2. Both subjects reported only mild symptoms. 3. SARS-CoV-2 Seroprevalence: Of 4,932 individuals tested for SARS-CoV-2 IgG antibodies, 64 subjects were categorized as seropositive, corresponding to a seroprevalence of 1.3% (95% confidence interval, 1.0 – 1.7%). Altogether, 19 children (0.8%; 95% confidence interval, 0.5 – 1.2%) and 45 parents (1.8%; 95% confidence interval, 1.3 – 2.4%) were seropositive. Seropositivity was found in 7/1,122 in the age group of 1 – 5 years (0.6%; 95% confidence interval, 0.3 – 1.3%), 12/1,358 in the age group of 6 – 10 years (0.9%, 95% confidence interval, 0.5 – 1.6%) and 45/2,468 in the parent group (1.8%; 95% confidence interval, 1.3 – 2.4%).

3 Study design and conduct This is a non-interventional, uncontrolled, open, national multi-center, cross-sectional study on the point prevalence of SARS-CoV-2 infections as determined by RT-PCR testing of nasopharyngeal and oropharyngeal swabs and presence of SARS-CoV-2 antibodies in serum (German Registry for Clinical Studies (DRKS), study ID 00021521). The four study centers are the University Children’s hospitals at Freiburg, Heidelberg, Tübingen and Ulm. The study was announced in national and respective local newspapers as well as in social nd th networks from April 22 to 30 , 2020. Subjects were investigated during the period from nd th April 22 to May 15 , 2020. Recruitment was through public announcement of the parent- child study and study participation upon application was random and voluntary. The study protocol was approved by the institutional review boards and the independent ethics committees of each center, and the study was conducted according to the Declaration of Helsinki. Written informed consent was obtained from all parents/guardians, with assent from children when appropriate for their age. Eligibility criteria and study procedure Subjects were eligible for enrollment if they met the following inclusion criteria: (i) Children (male or female) aged 1 to 10 years, (ii) one parent (male or female) without age limit, (iii) child and parent living in the same household, (iv) residency in the state of Baden- Württemberg, (v) written consent to the study had been obtained. Key exclusion criteria were (i) severe congenital diseases (e.g. infantile cerebral palsy, severe congenital malformations), (ii) congenital or acquired immunodeficiencies, (iii) laboratory-confirmed SARS-CoV-2 infection in the child or participating parent before study enrollment, (iv) lack of consent for child or parent. Study participants received a questionnaire (see digital Supplementary Material) on social status, occupation, age and chronic illnesses of the parents. Questions concerning the children included (i) chronic illnesses, (ii) attendance of day-care centers, kindergarten or elementary school, (iii) since when the children have been in home care or whether they have continued to attend day-care centers, kindergarten or after-school care within the framework of emergency child care. For both children and parents, we inquired (i) whether there had been contact to someone with laboratory-confirmed SARS-CoV-2 infection, (ii) whether SARS-CoV-2 infection had already been diagnosed in the participants themselves (exclusion criterion), or (iii) whether health problems (fever, cough, "common cold", diarrhea or loss of smell and taste) had occurred since the end of February 2020 (outbreak of the COVID-19 pandemic in Europe). Study objectives The following primary study objectives were predefined in the study protocol: (i) What is the rate of SARS-CoV-2 RNA positive children aged 1-10 years and one parent in a population- based sample in Baden-Württemberg? (ii) What is the seroprevalence of SARS-CoV-2 antibodies in the indicated study collectives? (iii) Are there any age-dependent subgroups among children aged 1-10 years with regard to different seroprevalence? In addition, the following secondary study objectives were predefined in the study protocol but not part of this interim analysis (exception iii): (i) What is the rate of intra-family transmission? (ii) What is the proportion of parents or children who have been diagnosed

4 with antibodies against SARS-CoV-2 but have developed few or no symptoms? (iii) Does the housing situation have an impact on the transmission rate between parent and child? (iv) Does the professional environment of the parents have an impact on the family risk of infection? (v) Does family size impact the family risk of infection? Laboratory analyses Sample collection and storage Specimens for PCR diagnostics were collected as oropharyngeal/nasopharyngeal swabs in e.g. eSwab, Copan, Milan, Italy or Sigma-Virocult, respectively. Collected samples were transported in sterile containers, delivered to the diagnostic laboratory within a few hours, and examined directly or stored at 4°C until further processing. Blood samples for serological analyses were centrifuged and serum was collected. Serum samples were either examined directly or stored at 4°C until further processing. Serum shipment to test centers at other sites was by overnight courier at room temperature. RNA isolation from nasal and oropharyngeal swabs and reverse-transcriptase polymerase chain reaction (RT-PCR) RNA was isolated from nasopharyngeal swabs using QIAGEN Kits (QIAGEN, Hilden, Germany) automated on the QIASymphony instrument (DSP Virus pathogen mini kits), and eluted in 115 μl elution buffer. Ten μl of extracted samples were used in a 20 μl RT-PCR reaction, carried out using various reagent mixes: LightMix Modular SARS and Wuhan CoV E-gene, LightMix Modular SARS and Wuhan CoV N-gene, LightMix Modular Wuhan CoV RdRP-gene and LightMix Modular EAV RNA Extraction Control (TIBMOLBIOL, Berlin, Germany) and LightCycler Multiplex RNA Virus Master (Roche, Mannheim, Germany) according to manufacturer's instructions. RT-PCR was performed on LightCycler 480 instruments (Roche, Mannheim, Germany). Thermal profile was as follows: reverse transcription step at 55 °C for 5 min, followed by denaturation at 95°C for 5 min, and 45 amplification cycles (denaturation at 95°C for 5 sec, annealing at 60°C for 15 sec, and elongation at 72°C for 15 sec). Serological analyses The CE certified diagnostic tests applied here for SARS-CoV-2 serological testing have a high specificity according to the manufacturer’s documentation (Euroimmun SARS-CoV-2 IgG ELISA: 99.3% validated on 1,153 samples; Roche Elecsys Anti-SARS-CoV-2: 99.81% validated on 10,533 samples; Mikrogen recomWell ELISA 98.7%). With specificities <100%, however, false positive results would make up a substantial proportion of all positive results in populations with a low seroprevalence when only a single test is used. To increase specificity, we therefore applied a combination of two or more serological test methods. All sera were separately tested for the presence of SARS-CoV-2 specific IgG antibodies by ELISA and by immunofluorescence (IF). Sera for which unclear or discordant results were obtained in the combination of these two assays were further assessed by ECLIA or a second ELISA and in some instances by an in-house Luminex based assay detecting antibodies against several SARS-CoV-2 proteins. Sera are classified as positive when they scored positive in both initial tests and negative when they scored negative in both initial tests. Sera with discordant results are classified as positive when they also tested positive in

5 one or more further tests detecting a different SARS-CoV-2 antigen, and are classified as negative otherwise. Sera classified as positive were subsequently tested for neutralizing titer (ongoing analysis). ELISA measurements for determination of reactivity against the S1 domain of the viral spike protein were carried out using the Euroimmun Anti-SARS-CoV-2-ELISA (IgG) (Euroimmun AG, Lübeck, Germany, EI 2606-9601 G) test kit according to the manufacturer’s protocol. Samples were processed on a Euroimmun Analyzer I instrument according to the manufacturer’s instructions. Measurements for determination of reactivity against the viral nucleocapsid (N) protein were carried out using either the Elecsys® Anti-SARS-CoV-2 test kit (#09 203 095 190, Roche Diagnostics, Mannheim) processed on a Roche Cobas 601 module (sera from Heidelberg, Tübingen and Ulm) or by recomWell SARS-CoV2 IgG ELISA (Mikrogen GmbH Martinsried, Germany) on a BEP III analyzer (sera from Freiburg). All measurements were performed according to the manufacturers´ instructions. SARS-CoV-2 specific antibodies were further determined using indirect immunofluorescence on sub-confluent VeroE6 African green monkey cells in 96-well plates infected or not with SARS-CoV-2 (BavPat1/2020 strain, European Virus Archive). Cells were fixed with paraformaldehyde and either permeabilized (sera from Heidelberg, Tübingen and Ulm) or not (Freiburg). Fluorescence labeled secondary antibody (Goat anti-human IgG-AlexaFluor 488, Invitogen, Thermofisher Scientific or anti-human IgG-Cy3, Jackson ImmunoResearch) was used for detection. Plates with sera from Freiburg were subjected to fluorescence microscopy using a Zeiss Observer.Z1 inverted epifluorescence microscope (Carl Zeiss) equipped with an AxioCamMR3 camera. Seropositivity was determined by visually comparing SARS-CoV-2-infected and uninfected cells incubated in parallel with the same person´s serum. Plates with sera from Heidelberg, Tübingen and Ulm were analyzed using a semi-quantitative, semi-automated procedure described in detail in a manuscript available as pre-publication text (Pape, Remme et al., https://www.biorxiv.org/content/10.1101/2020.06.15.152587v1). Nine fluorescence images per well were acquired with a motorized Nikon Ti2 widefield microscope, automatically segmented, feature-extracted, analyzed and classified as described in Pape, Remme et al. Determination of neutralizing antibody titers (NT) Neutralizing antibodies were determined by infection of subconfluent VeroE6 cells with SARS-CoV-2 incubated or not with different concentrations of participants´ sera. Serial two‐ fold dilutions of sera were prepared in OptiMEM medium or PBS and incubated with SARS‐ CoV‐2 for 1 h at 37°C or room temperature prior to infection of VeroR6 cells. For the analysis of sera from Heidelberg, Tübingen and Ulm, infection was scored at 20 h post infection by immunostaining of fixed cells using the anti ds-RNA mouse monoclonal J2 antibody (Scicons, 1:1000) and a secondary anti‐mouse HRP‐coupled antibody (Merck, Darmstadt, Germany, 1:1000). The signal was developed using KPL SureBlueTM 3,3',5,5'- tetramethylbenzidine peroxidase substrate (Seracare, Milford, MA, USA) for 5 min and stopped by the addition of 0.5 M sulfuric acid. Absorbance was measured on a Tecan Sunrise plate reader (Tecan, Männedorf, Switzerland) at 450 nm with reference wavelength 620 nm. Data were normalized to the no‐serum control (100%) and a mock‐infected control (0%). In order to ensure reproducibility of results, one serum sample from a healthy donor and one sample from a symptomatic Covid-19 patient, collected at day 39 after onset of symptoms, were used in all experiments. Inhibitory dilution 50 (ID50), defined as serum dilution resulting in 50% reduction of normalized signal, was determined using the nonlinear

6 regression function of GraphPad Prism software or a custom Excel template using the same four parameter dose response equation. For sera from Freiburg, virus incubated with sera or not was removed from VeroE6 cells after 1.5 h incubation at room temperature, and the cells were overlaid with 0.6% Oxoid-agar in DMEM, 20 mM HEPES (pH 7.4), 0.1% NaHCO3, 1% FBS and 0.01% DEAE-Dextran. Cells were formaldehyde fixed 48 h post-infection and stained with 1% crystal violet upon removal of the agar overlay. Plaque forming units (PFU) were counted manually. The number of plaques counted for serum-treated wells was compared to the average number of plaques in the untreated negative controls that were set to 100%. Statistical analysis Analyses were performed using R version 4.0.0 (R Core Team, 2020). Prevalence for seropositivity in different age groups were computed using the 'epiR' package (version 1.0- 14) using exact method according to Collett (1999, p. 24). Results for continuous variables are presented as median with interquartile range (IQR), unless stated otherwise. Results Study Population The entire study population comprised 5,042 subjects (2,521 children and 2,521 corresponding parents). The present interim analysis was performed in 4932 subjects (2,466 children and 2,466 corresponding parents), because data cleaning regarding missing or ambiguous values in the remaining subjects is still ongoing. Subject demographics of the study population for interim analysis (n=2,466 parent-child pairs) are given in Table 1. The region of residence of study subjects analyzed according to the three-digit postal code is depicted in Figure 1. The large majority of participants came from the study centers Freiburg, Heidelberg, Tübingen and Ulm or the adjacent regions, but the catchment area also included other regions of the federal state of Baden-Württemberg. Detection of acute SARS-CoV-2 infection by RT-PCR nd th Among the 4,932 persons who were tested by RT-PCR between April 22 and May 15 , 2020, only two subjects (0.04%), one child and the corresponding parent, were tested positive for SARS-CoV-2 RNA. Both subjects reported only mild symptoms. SARS-CoV-2 Seroprevalence Of 4,932 persons tested for SARS-CoV-2 IgG antibodies, 64 subjects were categorized as seropositive, corresponding to a seroprevalence of 1.3% (95% confidence interval, 1.0 – 1.7%). Altogether, 19 children (0.8%; 95% confidence interval, 0.5 – 1.2%) and 45 parents (1.8%; 95% confidence interval, 1.3 – 2.4%) were seropositive. The number of seropositive individuals in the age group 1 – 5 years was 7/ 1120 (0.6%; 95% confidence interval, 0.3 – 1.3%), in the age group of 6 – 10 years 12/ 1346 (0.9%, 95% confidence interval, 0.5 – 1.6%) and in the parent group 45/ 2466 (1.8%; 95% confidence interval, 1.3 – 2.4%). In 68% of seropositive children, the corresponding parent was also seropositive. Further statistical analysis is in progress.

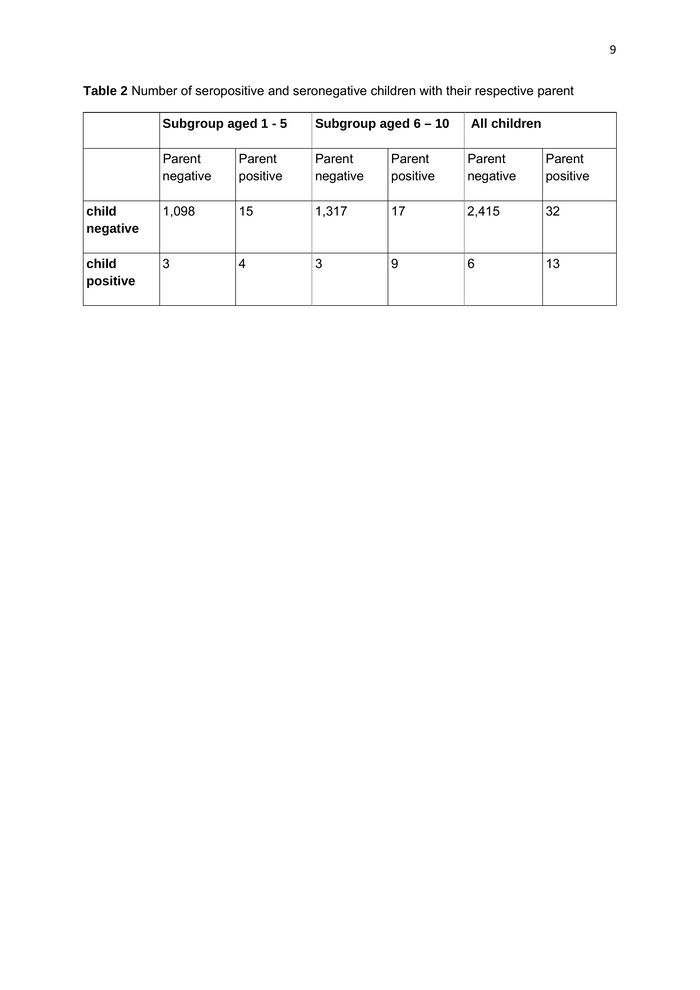
7 Table 2 shows the number of seropositive and seronegative children or parents within one family. Overall, there were 32 seropositive parents with a seronegative child and 6 seropositive children with a seronegative parent. In the subgroup aged 1-5 years, there were 15 seropositive parents with a seronegative child, and three seropositive children with a seronegative parent. For the subgroup aged 6 – 10 years, there were 17 seropositive parents with a seronegative child and 3 seropositive children with a seronegative parent (95% confidence interval, 1.6 – 30.2). The number of reported siblings in the families was n=0 in 462 families, n=1 in 1,412 families, n=2 in 466 families, n=3 in 96 families, n=4 in 14 families and n=5 in 2 families. In 14 families, information about siblings is missing. The association of the number of siblings with seropositivity of either a parent, a child or both will be investigated. 586 of the 2,466 children analyzed (23.7%) attended emergency child care facilities during the study period. Table 3 shows the association of the characteristic “emergency child care” with seropositivity. Further statistical analysis is ongoing. Of 2,466 parents, 235 (14 seropositive) reported contacts with COVID-19 patients, 2,209 (31 seropositive, 22 parents missing, but no seropositves) denied such a contact. Among all seropositive adults (n=45), five (11%) reported to work in the health care system (These are n=6: paramedic, n=1; medical doctor, n=2; caregiver, n=1; nurse, n=2).

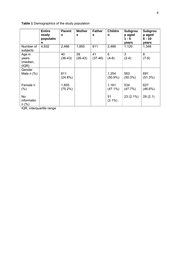
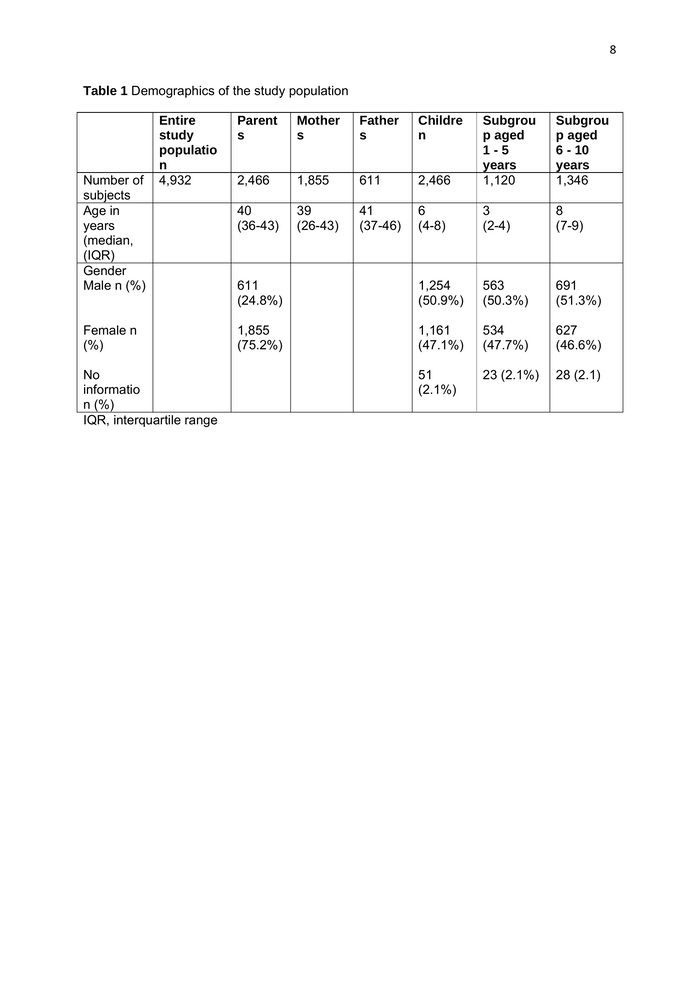
8 Table 1 Demographics of the study population Number of subjects Age in years (median, (IQR) Gender Male n (%) Entire study populatio n 4,932 Female n (%) No informatio n (%) IQR, interquartile range Parent s Mother s Father s Childre n 2,466 Subgrou p aged 1-5 years 1,120 Subgrou p aged 6 - 10 years 1,346 2,466 1,855 611 40 (36-43) 39 (26-43) 41 (37-46) 6 (4-8) 3 (2-4) 8 (7-9) 611 (24.8%) 1,254 (50.9%) 563 (50.3%) 691 (51.3%) 1,855 (75.2%) 1,161 (47.1%) 534 (47.7%) 627 (46.6%) 51 (2.1%) 23 (2.1%) 28 (2.1)
9 Table 2 Number of seropositive and seronegative children with their respective parent Subgroup aged 1 - 5 Subgroup aged 6 – 10 All children Parent negative Parent positive Parent negative Parent positive Parent negative Parent positive child negative 1,098 15 1,317 17 2,415 32 child positive 3 4 3 9 6 13
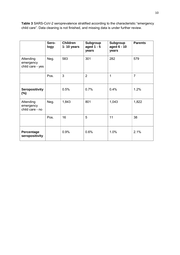
10 Table 3 SARS-CoV-2 seroprevalence stratified according to the characteristic “emergency child care”. Data cleaning is not finished, and missing data is under further review. Sero- logy Children 1- 10 years Subgroup aged 1 - 5 years Subgroup aged 6 - 10 years Parents 583 301 282 579 3 2 1 7 0.5% 0.7% 0.4% 1.2% Neg. 1,843 801 1,043 1,822 Pos. 16 5 11 38 0.9% 0.6% 1.0% 2.1% Attending Neg. emergency child care - yes Pos. Seropositivity (%) Attending emergency child care - no Percentage seropositivity